Editorial by Prof Nikolaos Papanikolaou, Scientific manager of the ProCAncer-I project
ProCAncer-I is a collaborative project that combines the efforts of 20 clinical and technical partners in Europe and US. The focus is to collect the biggest in size, diverse dataset called ProstateNET to develop AI models that can address open, unmet clinical questions in the field of prostate cancer. We decided to focus on prostate cancer, since it has a very high prevalence while the current methods and tools referring from early diagnosis to treatment selection and patient management, are not ideal resulting in overdiagnosis and overtreatment in patients with non-aggressive tumors and missed treatment opportunities for patients with aggressive disease.
Therefore, we wanted to incorporate cutting-edge technologies like AI and radiomics to develop models and validate them with the highest standards, so they may be used in the clinical practice and improve diagnostic and treatment outcomes. A total number of about 17000 prostate MRI studies accompanied with clinical data are projected to be available in the ProCAncer-I platform. These studies will undergo curation using AI powered tools in the platform to transform them into forms that can be used with machine learning methods to produce clinical value.
The retrospective data collection and uploading is critical to the success of ProCAncer-I, since it will be the backbone of ProstateNET providing a big dataset comprising of several millions of prostate representations visualized with different MRI contrasts (T2, DWI, ADC, DCE), coming from 11 geographically diverse clinical providers. In addition, the clinical institutions are regional private diagnostic centers, public hospitals, specialized anti-cancer centers as well as university hospitals that are reference centers for prostate cancer. Apart from the size of the data, an equally important quality of ProstateNET is the data diversity based on 1.5T and 3T scanners, with or without the use of an endorectal coil, and many different sequence parameter combinations, that brings the ProstateNET very close to real world data. The latter will have very positive effects towards developing not only high performance AI models, but also generalizable which at the moment is the drawback of existing AI models that are not performing as promised in each and every setup (hospital, scanner, protocol, patient cohort).
Following a very tedious standardization process related to data curation, we got managed to agree on the minimum necessary non-imaging clinical variables as well as the type of MRI images that should be present to make a patient eligible for uploading to ProCAncer-I platform. The data curation and uploading process has already started and it is planned to be finalized in month 24, when the necessary computational pipelines for training models will become available. So far about 30% of the projected data has been curated and uploaded to the platform, however, we already exceeded the 1.5M images kpi, since at the moment we are approaching 2M images (1.97M). The first wave of retrospective data are used to perform some proof of concept studies related to AI tasks (segmentation, detection, sequence identification, preprocessing pipelines, etc). We are in month 20 of the project, and we have already defined the strategy regarding the AI methods to be used. The types of AI or ML models that will be developed and validated under the scope of ProCAncer-i project can be grouped into detection, segmentation and classification models, and these are the main ones to support the 9 clinical use cases. 4 additional models will serve the need to enhance the quality of the data, including a sequence identification model, an image quality assessment model, an image denoising model and finally an image enhancement model. Starting with sequence identification, we have experienced very heterogeneous data at the sequence name level, meaning that for the same type of sequence you might encounter different sequence names. This can be explained in the basis of vendor related differences, institution naming strategies or even radiographer preferences since the sequence name is a free text variable that can be entered by the radiographer performing the relative MRI examination. The idea is to use several metadata available in our repository that are extracted from the dicom header of each series, map them to 5 different entities and train a ML model (fig 1).
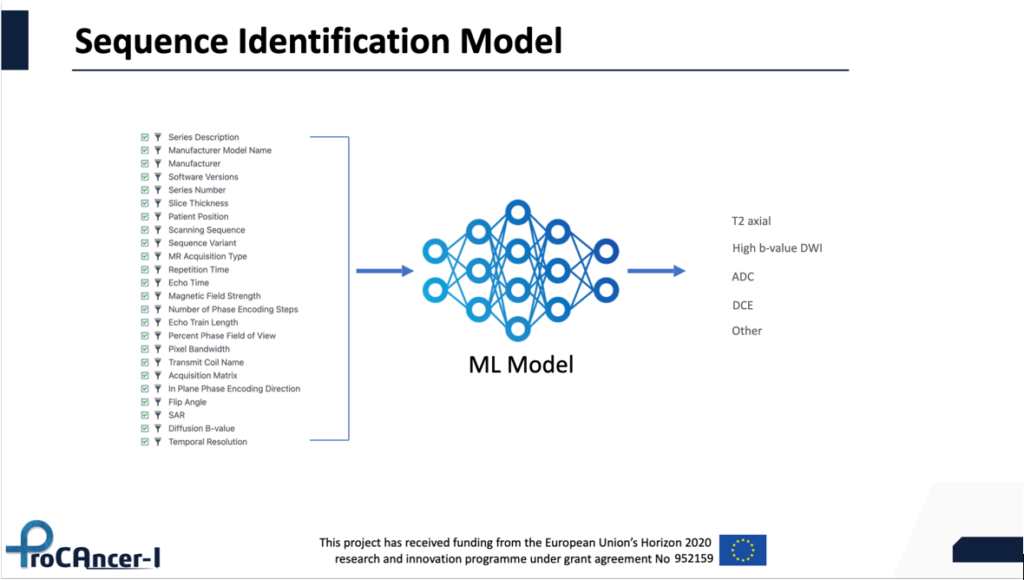
Fig 1. Sequence identification model idea
In this way, the necessary standardization at the sequence name level can be achieved automatically. Another AI model that we are considering is an IQ assessment model that will be trained using as an input the MRI images. In a subset of patients, a group of expert radiologists will provide rates of the image quality that will act as the ground truth to train a classifier. After training, the model will be deployed on the ProCAncer-I platform and predict the quality of each image that is uploaded prior to its transfer to the prostateNET repository. This model will guarantee that the repository is not contaminated by exams of unacceptable image quality (fig 2).
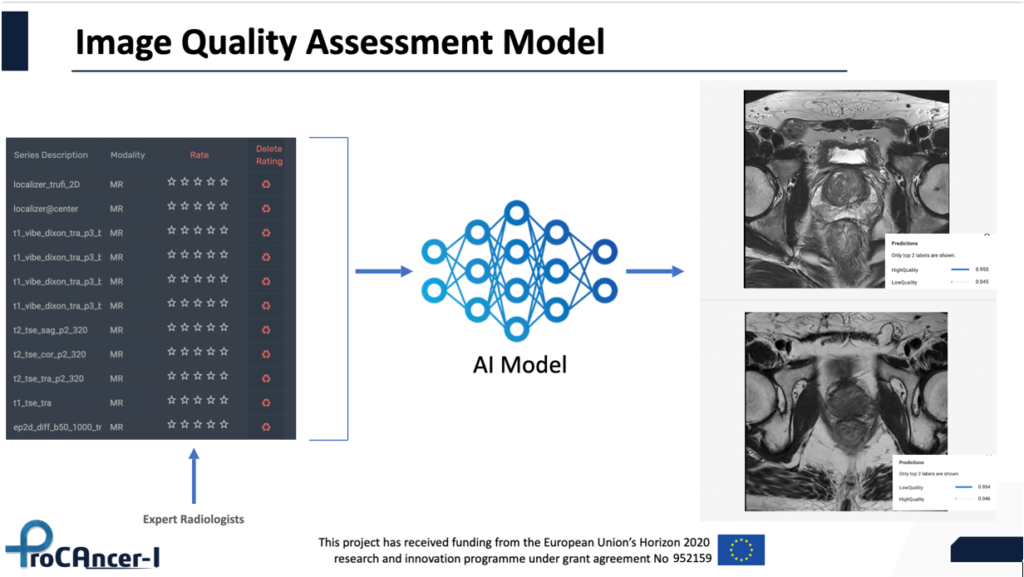
Fig 2. Image quality Assessment AI model
The main AI models that will address all clinical use cases, will be developed based on a three-stage strategy. Initially, we will train models for every UC, making use of the whole retrospective dataset, without making any exclusions vendor wise, protocol wise or field strength wise. These master models will be trained on diverse real-world data and will be used for benchmarking purposes, as well as to act as the backbone of the models at the second stage. More specifically, and through transfer learning, vendor specific models will be fine-tuned using 20% of vendor specific data of the prospective dataset using the master models as pre-trained architectures. In the third stage, vendor neutral models will be developed as meta-learners ensembling predictions from the three vendor specific models. Finally, we will pool the data into three nodes according to the vendor they have been acquired from and train vendor neutral models using distributed learning (fig 3).
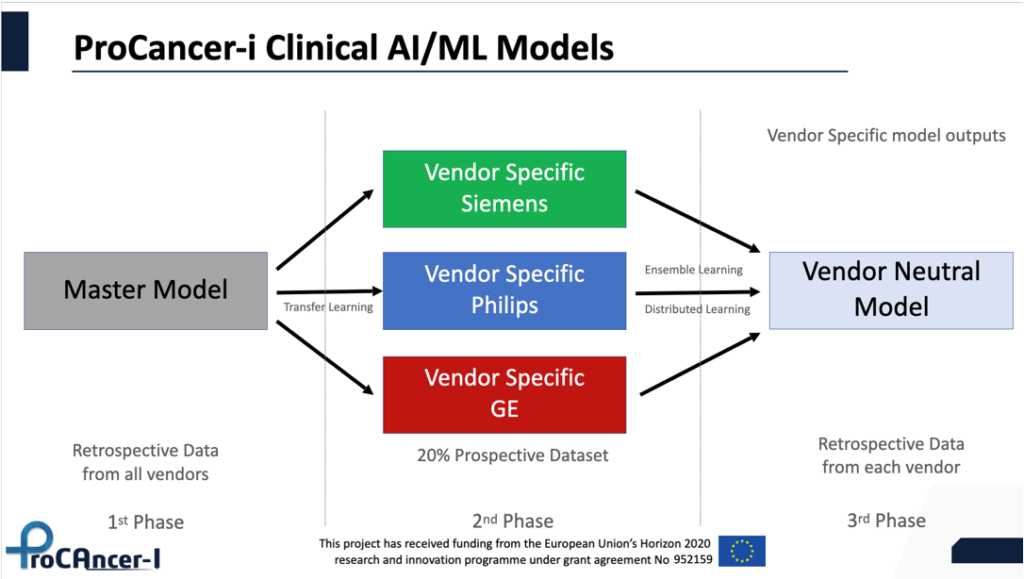
Fig 3. Clinical AI models of the ProCAncer-I project
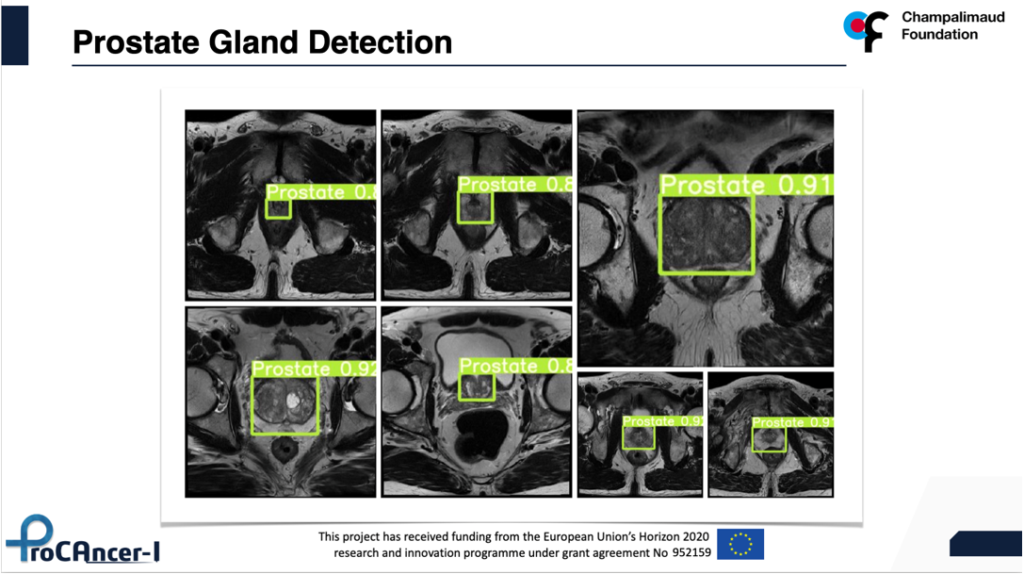
The detection models will be exclusively based on deep neural network architectures like YOLO5 to either detect the location of the whole prostate gland or the index lesion which is the biggest in size and the most aggressive. 5 models will be trained per category one master model, 3 vendor-specific and 1 vendor neutral model, leading to a total of 10 detection models.
In conclusion, the project is progressing fast and the initial results are very exciting and encouraging to continue building upon the ProCAncer-I platform, harnessing the power of high-quality curated data that we are producing, with the ultimate goal to provide to our end-users (clinicians and patients) reliable, explainable and trustworthy AI models that can create value and help individual patients as well as health care systems to improve clinical outcomes in prostate cancer.

